Legionella pneumophila
| Legionella pneumophila | |
|---|---|
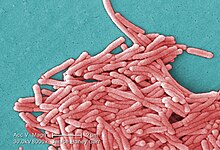
| |
| Colorized scanning electron micrograph image of L. pneumophila | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Pseudomonadota |
| Class: | Gammaproteobacteria |
| Order: | Legionellales |
| Family: | Legionellaceae |
| Genus: | Legionella |
| Species: | L. pneumophila
|
| Binomial name | |
| Legionella pneumophila Brenner DJ, Steigerwalt AG, McDade JE 1979
| |
Legionella pneumophila is an aerobic, pleomorphic, flagellated, non-spore-forming, Gram-negative bacterium.[1][2] There are fourteen serogroups of L. pneumophila. L. pneumophila is a facultative intracellular parasite that infects soil amoebae and freshwater amoeboflagellates for replication.[3][4] This pathogen is thus found commonly near freshwater environments[5] and invades the unicellular life, using them to carry out metabolic functions.[6] Due to L. pneumophila’s ability to thrive in water, it can grow in water filtration systems, leading to faucets, showers, and other fixtures and then spread through aerosolized water droplets.[6]
L. pneumophila is also the causative agent of Legionnaires' disease, also known as legionellosis.[4] Due to the nature of replication of the bacteria, it is able to reproduce freely in human macrophages.[4] Once infected, this bacterium can cause pneumonia with symptoms such as fever, delirium, diarrhea, and decreased liver and kidney function.[7] L. pneumophila infections can be diagnosed by a urine antigen test.[8][9] The infections caused by the bacteria can be treated with fluoroquinolones and azithromycin antibiotics.[8]
Characterization
[edit]L. pneumophila is a coccobacillus.[10] It is a Gram-negative, aerobic bacterium unable to hydrolyse gelatin or produce urease. It is also non-fermentative. L. pneumophila is neither pigmented nor does it autofluoresce. It is oxidase- and catalase-positive, and produces beta-lactamase.[11] L. pneumophila colony morphology is gray-white with a textured, cut-glass appearance; it also requires cysteine and iron to thrive.[12] It grows on buffered charcoal yeast extract agar as well as in moist environments, such as tap water, in "opal-like" colonies.[12]
The bacterium is facultative and able to persist in a range of environments. Due to a specialized characterization the bacterium may live intra- or extracellularly. This explains why L. pneumophila is present in many freshwater environments as well as the alveolar macrophages within the human lungs. This is achieved through its two forms: a transmissive and replicative form. Transition between the two occurs throughout its lifecycle; It is activated by changing availability of metabolic/nutritional resources in its environment. To infect its hosts the bacteria use their transmissive form. The replicative form then follows to carry out proliferation.[13]
Cell membrane structure
[edit]L. pneumophila is a Gram-negative organism based on the cell membrane structure, which is composed of two membranes separated by periplasmic space. Its unique outer membrane composed of lipoproteins, phospholipids, and other proteins is the distinguishing feature of Legionella spp. Like most gram-negative bacteria, L. pneumophila have a three part lipopolysaccharide. Legionella spp. possess unique lipopolysaccharides (LPS) extending from the outer membrane leaflet of the outer cell membrane that play a role in pathogenicity and adhesion to a host cell. Lipopolysaccharides are the leading surface antigen of all Legionella species including L. pneumophila. [14]
The bases for the somatic antigen specificity of this organism are located on the side chains of its cell wall. The chemical composition of these LPS side chains both with respect to components and arrangement of the different sugars, determines the nature of the somatic or O-antigenic determinants, which are important means of serologically classifying many Gram-negative bacteria. L. pneumophila exhibits distinct chemical characteristics in its LPS structure that distinguish it from other Gram-negative bacteria. [15] The unique attributes are key factors in its serological identity and biological function. [15]
Ecology
[edit]
L. pneumophila is a bacterium that can be found in various environmental conditions. It can preside in temperatures ranging from 0-63 °C, a pH range of 5.0-8.5, and in dissolved oxygen levels of 0.2-15.0 mg/liter.[5]
It can be found in freshwater environments with a wide range of temperatures. Although it can be found in this wider range, it only multiplies within a temperature range of 25 and 42 °C.[5] With their ability to reside in water, they can also resist chlorination of water and pass into water control systems easily.[5] With this ability to infiltrate water systems, they can form biofilms in the walls of pipes which can lead to this bacterium being aerosolized through faucets, showers, sprinklers, and other fixtures, leading to infection after prolonged exposure.[6] The main source of L. pneumophila contamination is the water supply network and this has allowed L. pneumophila to grow and proliferate in places such as cooling towers, water systems of hospitals, hotels, and cruise ships.[5]
As a facultative intracellular parasite, L. pneumophila can invade and replicate inside protozoa in the environment, especially within the species of the genera Acanthamoeba and Naegleria, which can thus serve as a reservoir for L. pneumophila. These hosts will then provide protection against unfavorable physical and chemical conditions, such as chlorination.[3]
Biofilms
[edit]Biofilms are specialized, surface attachment communities that can consist of one microbe, or multiple different ones, ranging from bacteria, algae, and protozoa.[5] These protective matrixes enable the microbe to live for extended periods of time in low-nutrient environments.[11] The bffA gene's key purpose is to regulate biofilm formation. [16] Biofilms on plumbing systems and in water distribution systems is where a lot of L. pneumophila can survive.[5] Between 2009 and 2010, L. pneumophila contributed to 58% of the total waterborne disease outbreaks associated with drinking water in the United States, causing an increase in research surrounding L. pneumophila biofilms and proliferation.[17] Material also plays a role in biofilm proliferation. In water piping, L. pneumophila was more commonly found in plastic pipes at 40 °C, rather than a copper pipe, which actually inhibited growth.[5]
Environmental protozoa
[edit]Legionella is a genus that is known to infect and multiply within species of environmental protists. We know of at least 20 different species of protozoa that support the intracellular replication of L. pneumophila. [18] This bacterium can infect and survive within protozoa genera such as Acanthamoeba, Vermamoeba and Naegleria. The bacteria are surrounded by an resistant cyst while residing inside the host cell, allowing them to survive harsh environmental conditions such as chlorine, which is commonly used in water treatment systems.[19]
Although it is known that protozoa play an important role in the ecology of L. pneumophila, there is little data on how these organisms interact with other microbes in the environment, and how this affects L. pneumophila overall.[18]
Prevalence
[edit]L. pneumophila is the primary (>90%) causative organism for Legionnaires disease.[20] Roughly 2 per 100,000 people are infected each year in the EU.[21] L. pneumophila often infects individuals through poor quality water sources. Approximately 20% of reported Legionnaires disease cases come from healthcare, senior living, or travel facilities that have been exposed to water contaminated with L. pneumophila. [20] There may also be an increased risk of contracting L. pneumophila from private wells, as they are often unregulated and not as rigorously disinfected as municipal water systems.[22] Several large outbreaks of Legionnaire's Disease have come from public hot tubs due to the temperature range of the water being ideal for Legionella growth.[23][24]
Pathogenesis
[edit]

L. pneumophila's natural host is freshwater protozoa, but the bacteria is able to invade and replicate within human alveolar macrophages.[26] The internalization of the bacteria can be enhanced by the presence of antibody and complement, but is not absolutely required.[27] Internalization of the bacteria appears to occur through phagocytosis or coiling phagocytosis and is reliant on Dot/Icm type 4B secretion system (T4BSS).[26] Once internalized, the Dot/Icm system begins secreting bacterial effector proteins that recruit host factors to the Legionella containing vacuole (LCV). This process prevents the LCV from fusing with the lysosomes that would otherwise degrade the bacteria. Vesicles of the host cell's rough endoplasmic reticulum are attracted to the LCV, and these vacuoles supple the LCV with necessary lipids and proteins.[26] LCV membrane integrity requires a steady supply of host lipids, such as cellular cholesterol and the cis-monounsaturated fatty acid, palmitoleic acid.[28][29] L. pneumophila replication occurs within the LCV. Once nutrients are depleted, the bacteria gain flagella and cytoxicity. To exit the host cell, L. pneumophila lyses the LCV and resides in the cytoplasm. In the cytoplasm, L. pneumophila inhibit organelle and plasma membrane function and structure which ultimately leads to osmotic lysis of the host cell.[30]
Virulence factors
[edit]L. pneumophila exhibits a unique lipopolysaccharide (LPS) structure that is highly hydrophobic due to its being densely packed with branched fatty acids, and elevated levels of O-acetyl and N-acetyl groups.[31] This structure helps prevent interaction with a common LPS immune system co-receptor, CD14.[31] There is also a correlation between an LPS with a high molecular-weight and the inhibition of phagosome-lysosome fusion.[31] L. pneumophila produces pili of varying lengths. The two pili proteins: PilE and Prepilin peptidase (PilD) are responsible for the production of type IV pili and subsequently intracellular proliferation.[32] L. pneumophila possesses a singular, polar flagellum that is used for cell motility, adhesion, host invasion, and biofilm formation.[31] The same regulators that control flagellation also control lysosome avoidance and cytotoxicity.[31] The macrophage infectivity potentiator is another key component of host cell invasion and intracellular replication. MIP displays peptidyl–prolyl cis/trans isomerase (PPIase) activity which is crucial for survival within the macrophage, along with transmigration across the lung epithelial barrier.[31][32]
Another key virulence factor of L. pneumophila is iron acquisition, the microbe has two methods of iron uptake that it utilizes. Ferrous iron is collected through the use of a transport system involving an inner-membrane protein known as protein FeoB. Optimal intracellular infection is achieved in amoebae and macrophages via this transport system. The second form of uptake, involving ferric iron, is achieved through an iron chelator known as legiobactin. This is secreted by L. pneumophila when the microbes is being grown in a low iron chemically designed media. [33]
Dot/Icm type IV secretion system
[edit]The bacteria use a type IVB secretion system known as Dot/Icm to inject effector proteins into the host. These effectors are involved in increasing the bacteria's ability to survive inside the host cell. L. pneumophila encodes for over 330 "effector" proteins,[34] which are secreted by the Dot/Icm translocation system to interfere with host cell processes to aid bacterial survival. It has been predicted that the genus Legionella encodes more than 10,000 and possibly up to ~18,000 effectors that have a high probability to be secreted into their host cells.[35][36]
One main way in which L. pneumophila uses its effector proteins is to interfere with fusion of the Legionella-containing vacuole with the host's endosomes, and thus protect against lysis.[37] Knock-out studies of Dot/Icm translocated effectors indicate that they are vital for the intracellular survival of the bacterium, but many individual effector proteins are thought to function redundantly, in that single-effector knock-outs rarely impede intracellular survival. This high number of translocated effector proteins and their redundancy is likely a result of the bacterium having evolved in many different protozoan hosts.[38]
Legionella-containing vacuole
[edit]
For Legionella to survive within macrophages and protozoa, it must create a specialized compartment known as the Legionella-containing vacuole (LCV).[39] Through the action of the Dot/Icm secretion system, the bacteria are able to prevent degradation by the normal endosomal trafficking pathway and instead replicate. Shortly after internalization, the bacteria specifically recruit endoplasmic reticulum-derived vesicles and mitochondria to the LCV while preventing the recruitment of endosomal markers such as Rab5a and Rab7a. Formation and maintenance of the vacuoles are crucial for pathogenesis; bacteria lacking the Dot/Icm secretion system are not pathogenic and cannot replicate within cells, while deletion of the Dot/Icm effector SdhA results in destabilization of the vacuolar membrane and no bacterial replication.[40][41]
Metabolism
[edit]L. pneumophila uses glycolysis, the Entner-Doudoroff (ED) pathway, the pentose phosphate pathway (PP), and the citric acid cycle (TCA).[42] Although L. pneumophila can also perform gluconeogenesis, it does not have the genes to encode for 1,6-biphosphatases. Therefore, other enzymes are used to complete gluconeogenesis. One enzyme used instead is fructose 6-phosphate aldolase.[42] This trend is also present when it comes to the PP pathway which can occur without substrates such as 6-phosphogluconate dehydrogenase.[42] The ED and PP pathways are the main pathways for glucose metabolism in this organism. Along with these pathways, serine was found to be a major nutrient due to its ability to be turned into pyruvate, which is an important intermediate in metabolic pathways in L. pneumophila.[42]
Although glucose metabolism is used, it is not one of the main synthesis pathways within the organism. While using media containing glucose, growth of L. pneumophila did not increase and carbohydrates were not considered an important carbon source within L. pneumophila. Glucose can act as a co-substrate only under certain conditions, as this microbe uses amino acids more frequently and efficiently.[42]
Nutrient acquisition
[edit]Legionella is auxotrophic for seven amino acids: cysteine, leucine, methionine, valine, threonine, isoleucine, and arginine. Once inside the host cell, Legionella needs nutrients to grow and reproduce. Inside the vacuole, nutrient availability is low; the high demand of amino acids is not covered by the transport of free amino acids found in the host cytoplasm. To improve the availability of amino acids, the parasite promotes the host mechanisms of proteasomal degradation. This process in L. pneumophila includes the SCF1 ubiquitin ligase and the AnkB F-Box effector, which is farnesylated by the activity of three host enzymes localized in the membrane of the LCV: farnesyltransferase, Ras-converting enzyme-1 protease, and ICMT. Farnesylation allows AnkB to get anchored into the cytoplasmic side of the vacuole. SCF1 and AnkB interact with each other to degrade Lys-linked polyubiquitinated proteins.[43] This generates an excess of free amino acids in the cytoplasm of L. pneumophila-infected cells that can be used for intravacuolar proliferation of the parasite.
The K48-linked polyubiquitination is a marker for proteasomal degradation that releases 2 to 24-amino-acid-long peptides, which are quickly degraded to amino acids by various oligopeptidases and aminopeptidases present in the cytoplasm. Amino acids are imported into the LCV through various amino acid transporters such as the neutral amino acid transporter B(0).[43]
The amino acids are the primary carbon and energy source of L. pneumophila, that have almost 12 classes of ABC-transporters, amino acid permeases, and many proteases, to exploit it. The imported amino acids are used by L. pneumophila to generate energy through the TCA cycle (Krebs cycle) and as sources of carbon and nitrogen. Because the amino acid degradation acts as the main carbon source for L. pneumophila, this microbe does not rely as heavily on glucose. Despite this, L. pneumophila does contain multiple amylases, such as LamB, which hydrolyzes polysaccharides into glucose monomers for metabolism. The loss of LamB can result in severe growth issues for L. pneumophila.[44]
However, promotion of proteasomal degradation for the obtention of amino acids and the hydrolyzation of polysaccharides may not be the only virulence strategies to obtain carbon and energy sources from the host. Type II–secreted degradative enzymes may provide an additional strategy to generate carbon and energy sources.[45] L. pneumophila is the only known intracellular pathogen to have a Type II Secretion System (secretome). In Type II Secretion, proteins are first translocated across the inner membrane into the periplasmic space. This process is mediated by either the Sec or Tat pathway. Soon after, the same proteins are then transported through a specific pore in the outer membrane to the exterior of the cell. This secretome is believed to have as many as 60 proteins incorporated into the system.[45]
Genomics
[edit]| NCBI genome ID | 416 |
|---|---|
| Ploidy | haploid |
| Genome size | 3.44 Mb |
| Number of chromosomes | 1 |
| Year of completion | 2004 |
There are 14 known serogroups of L. pneumophila but serogroup 1 is most commonly the causative agent of Legionnaires’ disease.[46] Three strains, L. pneumophila Philadelphia, L. pneumophila Paris, and L. pneumophila Lens, were isolated in 2004 which paved the way for understanding the molecular biology of the bacteria.[47] Subspecies, which are commonly defined by geographical location, share about 80% of their genome with variation between strains that account for the difference in virulence between subspecies.[47] The genome is relatively large of about 3.5 mega base pairs (mbp) which reflects a higher number of genes, corresponding with the ability of Legionella to adapt to different hosts and environments.[47] There is a relatively high abundance of eukaryotic-like proteins (ELPs). ELPs are beneficial for mimicking the bacteria' eukaryotic hosts for pathogenicity.[47] Other genes of L. pneumophila encode for Legionella-specific vacuoles, efflux transporters, ankyrin-repeat proteins, and many other virulence related characteristics.[47] In-depth comparative genome analysis using DNA arrays to study the gene content of 180 Legionella strains revealed high genomic plasticity and frequent horizontal gene transfer events.[47] Horizontal gene transfer allows L. pneumophila to evolve at a rapid pace and commonly is associated with drug resistance.[48]
Genetic transformation
[edit]Transformation is a bacterial adaptation involving the transfer of DNA from one bacterium to another through the surrounding liquid medium. Transformation is a bacterial form of sexual reproduction.[49] In order for a bacterium to bind, take up, and recombine exogenous DNA into its chromosome, it must enter a special physiological state referred to as "competence".
To determine which molecules may induce competence in L. pneumophila, 64 toxic molecules were tested.[50] Only six of these molecules, all DNA-damaging agents, caused strong induction of competence. These were mitomycin C (which introduces DNA inter-strand crosslinks), norfloxacin, ofloxacin, and nalidixic acid (inhibitors of DNA gyrase that cause double-strand breaks), bicyclomycin (causes double-strand breaks), and hydroxyurea (causes oxidation of DNA bases). These results suggest that competence for transformation in L. pneumophila evolved as a response to DNA damage.[50] Perhaps induction of competence provides a survival advantage in a natural host, as occurs with other pathogenic bacteria.[49]
Drug targets
[edit]Several enzymes in the bacteria have been proposed as tentative drug targets. For example, enzymes in the iron uptake pathway have been suggested as important drug targets.[33] Further, a cN-II class of IMP/GMP specific 5´-nucleotidase which has been extensively characterized kinetically. The tetrameric enzyme shows aspects of positive homotropic cooperativity, substrate activation and presents a unique allosteric site that can be targeted to design effective drugs against the enzyme and thus, the organism. Moreover, the enzyme is distinct from its human counterpart making it an attractive target for drug development.
Detection and treatment
[edit]Antisera have been used both for slide agglutination studies and for direct detection of bacteria in tissues using immunofluorescence via fluorescent-labelled antibodies. Specific antibodies in patients can be determined by the indirect fluorescent antibody test. ELISA and microagglutination tests have also been successfully applied.[9] A consistent method that has been used to detect the disease is the urine antigen test.[8]
Effective antibiotic treatment for Legionella pneumonia includes fluoroquinolones (levofloxacin or moxifloxacin) or, alternately, azithromycin.[8] There has been no significant difference found between using a fluoroquinolone or azithromycin to treat Legionella pneumonia.[8] Combination treatments with rifampicin are being tested as a response to antibiotic resistance during mono-treatments, though its effectiveness remains uncertain.[8]
These antibiotics work best because L. pneumophila is an intracellular pathogen.[51] Levofloxacin and azithromycin have great intracellular activity and are able to penetrate into Legionella-infected cells. The Infectious Diseases Society of America recommends 5–10 days of treatment with levofloxacin or 3–5 days of treatment with azithromycin; however, patients that are immunocompromised or have a severe disease may require an extended course of treatment.[51]
References
[edit]- ^ Madigan M, Martinko J, eds. (2005). Brock Biology of Microorganisms (11th ed.). Prentice Hall. ISBN 0-13-144329-1.
- ^ Heuner K, Swanson M, eds. (2008). Legionella: Molecular Microbiology. Caister Academic Press. ISBN 978-1-904455-26-4.
- ^ a b Rowbotham TJ (December 1980). "Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae". Journal of Clinical Pathology. 33 (12): 1179–1183. doi:10.1136/jcp.33.12.1179. PMC 1146371. PMID 7451664.
- ^ a b c Winn Jr., Washington C. (1996). Baron, Samuel (ed.). Medical Microbiology (4th ed.). University of Texas: The University of Texas Medical Branch at Galveston. ISBN 0-9631172-1-1.
- ^ a b c d e f g h Uzel A, Hames-Kocabas EE (2010). Legionella Pneumophila: From Environment to Disease. NOVA Science Publishers. pp. 5–8. hdl:11454/19520. ISBN 978-1-60876-947-6.
- ^ a b c Abdel-Nour M, Duncan C, Low DE, Guyard C (October 2013). "Biofilms: the stronghold of Legionella pneumophila". International Journal of Molecular Sciences. 14 (11): 21660–21675. doi:10.3390/ijms141121660. PMC 3856027. PMID 24185913.
- ^ Djordjevic Z, Folic M, Petrovic I, Zornic S, Stojkovic A, Miljanovic A, et al. (May 2022). "An outbreak of Legionnaires' disease in newborns in Serbia". Paediatrics and International Child Health. 42 (2): 59–66. doi:10.1080/20469047.2022.2108672. PMID 35944175. S2CID 251468797.
- ^ a b c d e f Viasus D, Gaia V, Manzur-Barbur C, Carratalà J (June 2022). "Legionnaires' Disease: Update on Diagnosis and Treatment". Infectious Diseases and Therapy. 11 (3): 973–986. doi:10.1007/s40121-022-00635-7. PMC 9124264. PMID 35505000.
- ^ a b Conway de Macario E, Macario AJ, Wolin MJ (January 1982). "Specific antisera and immunological procedures for characterization of methanogenic bacteria". Journal of Bacteriology. 149 (1): 320–328. doi:10.1128/jb.149.1.320-328.1982. PMC 216625. PMID 6172417.
- ^ Gomez-Valero, L.; Rusniok, C.; Buchrieser, C. (2009-09-01). "Legionella pneumophila: Population genetics, phylogeny and genomics". Infection, Genetics and Evolution. 9 (5): 727–739. Bibcode:2009InfGE...9..727G. doi:10.1016/j.meegid.2009.05.004. ISSN 1567-1348. PMID 19450709.
- ^ a b Iliadi, Valeria; Staykova, Jeni; Iliadis, Sergios; Konstantinidou, Ina; Sivykh, Polina; Romanidou, Gioulia; Vardikov, Daniil F.; Cassimos, Dimitrios; Konstantinidis, Theocharis G. (18 October 2022). "Legionella pneumophila: The Journey from the Environment to the Blood". Journal of Clinical Medicine. 11 (20): 6126. doi:10.3390/jcm11206126. ISSN 2077-0383. PMC 9605555. PMID 36294446.
- ^ a b Murdoch DR (January 2003). "Diagnosis of Legionella infection". Clinical Infectious Diseases. 36 (1): 64–9. doi:10.1086/345529. PMID 12491204.
- ^ Fonseca MV, Swanson MS (2014). "Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila". Frontiers in Cellular and Infection Microbiology. 4: 12. doi:10.3389/fcimb.2014.00012. PMC 3920079. PMID 24575391.
- ^ Iliadi, Valeria; Staykova, Jeni; Iliadis, Sergios; Konstantinidou, Ina; Sivykh, Polina; Romanidou, Gioulia; Vardikov, Daniil F.; Cassimos, Dimitrios; Konstantinidis, Theocharis G. (January 2022). "Legionella pneumophila: The Journey from the Environment to the Blood". Journal of Clinical Medicine. 11 (20): 6126. doi:10.3390/jcm11206126. ISSN 2077-0383. PMC 9605555. PMID 36294446.
- ^ a b Petzold, Markus; Thürmer, Alexander; Menzel, Susan; Mouton, Johan W; Heuner, Klaus; Lück, Christian (December 2013). "A structural comparison of lipopolysaccharide biosynthesis loci of Legionella pneumophila serogroup 1 strains". BMC Microbiology. 13 (1): 198. doi:10.1186/1471-2180-13-198. ISSN 1471-2180. PMC 3766260. PMID 24069939.
- ^ Marin, Courtney; Kumova, Ogan K.; Ninio, Shira (27 January 2022). "Characterization of a Novel Regulator of Biofilm Formation in the Pathogen Legionella pneumophila". Biomolecules. 12 (2): 225. doi:10.3390/biom12020225. ISSN 2218-273X. PMC 8961574. PMID 35204726.
- ^ Shen Y, Monroy GL, Derlon N, Janjaroen D, Huang C, Morgenroth E, et al. (April 2015). "Role of biofilm roughness and hydrodynamic conditions in Legionella pneumophila adhesion to and detachment from simulated drinking water biofilms". Environmental Science & Technology. 49 (7): 4274–4282. Bibcode:2015EnST...49.4274S. doi:10.1021/es505842v. PMC 4472476. PMID 25699403.
- ^ a b Dey R, Mameri MR, Trajkovic-Bodennec S, Bodennec J, Pernin P (September 2020). "Impact of inter-amoebic phagocytosis on the L. pneumophila growth". FEMS Microbiology Letters. 367 (18). doi:10.1093/femsle/fnaa147. PMID 32860684.
- ^ Muchesa P, Leifels M, Jurzik L, Barnard TG, Bartie C (2018). "Detection of amoeba-associated Legionella pneumophila in hospital water networks of Johannesburg". Southern African Journal of Infectious Diseases. 33 (3): 72–75. doi:10.1080/23120053.2018.1434060. S2CID 90027194 – via SAJID.
- ^ a b Donohue MJ, Pham M, Brown S, Easwaran KM, Vesper S, Mistry JH (June 2023). "Water quality influences Legionella pneumophila determination". Water Research. 238: 119989. Bibcode:2023WatRe.23819989D. doi:10.1016/j.watres.2023.119989. PMC 10351031. PMID 37137207.
- ^ Lupia T, Corcione S, Shbaklo N, Rizzello B, De Benedetto I, Concialdi E, et al. (February 2023). "Legionella pneumophila Infections during a 7-Year Retrospective Analysis (2016-2022): Epidemiological, Clinical Features and Outcomes in Patients with Legionnaires' Disease". Microorganisms. 11 (2): 498. doi:10.3390/microorganisms11020498. PMC 9965988. PMID 36838463.
- ^ Mapili K, Pieper KJ, Dai D, Pruden A, Edwards MA, Tang M, Rhoads WJ (April 2020). "Legionella pneumophila occurrence in drinking water supplied by private wells". Letters in Applied Microbiology. 70 (4): 232–240. doi:10.1111/lam.13273. PMID 31904109. S2CID 209894300.
- ^ Kuroki T, Amemura-Maekawa J, Ohya H, Furukawa I, Suzuki M, Masaoka T, et al. (February 2017). "Outbreak of Legionnaire's Disease Caused by Legionella pneumophila Serogroups 1 and 13". Emerging Infectious Diseases. 23 (2): 349–351. doi:10.3201/eid2302.161012. PMC 5324795. PMID 28098535.
- ^ Donovan CV, MacFarquhar JK, Wilson E, Sredl M, Tanz LJ, Mullendore J, et al. (March 2023). "Legionnaires' Disease Outbreak Associated With a Hot Tub Display at the North Carolina Mountain State Fair, September 2019". Public Health Reports. 139 (1): 79–87. doi:10.1177/00333549231159159. PMC 10905752. PMID 36971250. S2CID 257765345.
- ^ Pathology, Dr Yale Rosen Atlas of Pulmonary (2009-08-01), Legionella pneumonia, retrieved 2024-10-19
- ^ a b c Oliva, Giulia; Sahr, Tobias; Buchrieser, Carmen (2018-01-19). "The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism". Frontiers in Cellular and Infection Microbiology. 8: 3. doi:10.3389/fcimb.2018.00003. ISSN 2235-2988. PMC 5780407. PMID 29404281.
- ^ Rittig, M G; Krause, A; Häupl, T; Schaible, U E; Modolell, M; Kramer, M D; Lütjen-Drecoll, E; Simon, M M; Burmester, G R (October 1992). "Coiling phagocytosis is the preferential phagocytic mechanism for Borrelia burgdorferi". Infection and Immunity. 60 (10): 4205–4212. doi:10.1128/iai.60.10.4205-4212.1992. ISSN 0019-9567. PMC 257454. PMID 1398932.
- ^ Ondari, Edna; Wilkins, Ashley; Latimer, Brian; Dragoi, Ana-Maria; Ivanov, Stanimir S. (2023-01-02). "Cellular cholesterol licenses Legionella pneumophila intracellular replication in macrophages". Microbial Cell. 10 (1): 1–17. doi:10.15698/mic2023.01.789. PMC 9806796. PMID 36636491.
- ^ Wilkins, Ashley A.; Schwarz, Benjamin; Torres-Escobar, Ascencion; Castore, Reneau; Landry, Layne; Latimer, Brian; Bohrnsen, Eric; Bosio, Catharine M.; Dragoi, Ana-Maria; Ivanov, Stanimir S. (2024-02-12). "The intracellular growth of the vacuolar pathogen Legionella pneumophila is dependent on the acyl chain composition of host membranes". Frontiers in Bacteriology. 3. doi:10.3389/fbrio.2024.1322138. ISSN 2813-6144.
- ^ Newton, Hayley J.; Ang, Desmond K. Y.; van Driel, Ian R.; Hartland, Elizabeth L. (April 2010). "Molecular Pathogenesis of Infections Caused by Legionella pneumophila". Clinical Microbiology Reviews. 23 (2): 274–298. doi:10.1128/CMR.00052-09. ISSN 0893-8512. PMC 2863363. PMID 20375353.
- ^ a b c d e f Shevchuk O, Jäger J, Steinert M (2011). "Virulence properties of the legionella pneumophila cell envelope". Frontiers in Microbiology. 2: 74. doi:10.3389/fmicb.2011.00074. PMC 3129009. PMID 21747794.
- ^ a b Talapko J, Frauenheim E, Juzbašić M, Tomas M, Matić S, Jukić M, et al. (January 2022). "Legionella pneumophila-Virulence Factors and the Possibility of Infection in Dental Practice". Microorganisms. 10 (2): 255. doi:10.3390/microorganisms10020255. PMC 8879694. PMID 35208710.
- ^ a b Cianciotto NP (May 2015). "An update on iron acquisition by Legionella pneumophila: new pathways for siderophore uptake and ferric iron reduction". Future Microbiology. 10 (5): 841–851. doi:10.2217/fmb.15.21. PMC 4461365. PMID 26000653.
- ^ Ensminger AW (February 2016). "Legionella pneumophila, armed to the hilt: justifying the largest arsenal of effectors in the bacterial world". Current Opinion in Microbiology. 29: 74–80. doi:10.1016/j.mib.2015.11.002. PMID 26709975.
- ^ Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert JA, et al. (February 2016). "Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires". Nature Genetics. 48 (2): 167–175. doi:10.1038/ng.3481. PMC 5050043. PMID 26752266.
- ^ Gomez-Valero L, Rusniok C, Carson D, Mondino S, Pérez-Cobas AE, Rolando M, et al. (February 2019). "More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells". Proceedings of the National Academy of Sciences of the United States of America. 116 (6): 2265–2273. Bibcode:2019PNAS..116.2265G. doi:10.1073/pnas.1808016116. PMC 6369783. PMID 30659146.
- ^ Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR (June 2008). "Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors". Science. 320 (5883): 1651–1654. Bibcode:2008Sci...320.1651P. doi:10.1126/science.1158160. PMC 2514061. PMID 18566289.
- ^ Jules M, Buchrieser C (June 2007). "Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics". FEBS Letters. 581 (15): 2829–2838. Bibcode:2007FEBSL.581.2829J. doi:10.1016/j.febslet.2007.05.026. PMID 17531986. S2CID 23203471.
- ^ Isberg, O’Conner, Heidtman, Ralph, Tamara, Matthew (2008). "The Legionella pneumophila replication vacuole: making a cozy niche inside host cells". Nature Reviews. Microbiology. 7 (1): 13–24. doi:10.1038/nrmicro1967. PMC 2631402. PMID 19011659.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Harding CR, Stoneham CA, Schuelein R, Newton H, Oates CV, Hartland EL, et al. (July 2013). "The Dot/Icm effector SdhA is necessary for virulence of Legionella pneumophila in Galleria mellonella and A/J mice". Infection and Immunity. 81 (7): 2598–2605. doi:10.1128/IAI.00296-13. PMC 3697626. PMID 23649096.
- ^ Creasey EA, Isberg RR (February 2012). "The protein SdhA maintains the integrity of the Legionella-containing vacuole". Proceedings of the National Academy of Sciences of the United States of America. 109 (9): 3481–3486. doi:10.1073/pnas.1121286109. PMC 3295292. PMID 22308473.
- ^ a b c d e Eisenreich W, Heuner K (November 2016). "The life stage-specific pathometabolism of Legionella pneumophila". FEBS Letters. 590 (21): 3868–3886. doi:10.1002/1873-3468.12326. PMID 27455397. S2CID 8187321.
- ^ a b Price CT, Richards AM, Abu Kwaik Y (2014-08-26). "Nutrient generation and retrieval from the host cell cytosol by intra-vacuolar Legionella pneumophila". Frontiers in Cellular and Infection Microbiology. 4: 111. doi:10.3389/fcimb.2014.00111. PMC 4143614. PMID 25207263.
- ^ Best A, Price C, Ozanic M, Santic M, Jones S, Abu Kwaik Y (April 2018). "A Legionella pneumophila amylase is essential for intracellular replication in human macrophages and amoebae". Scientific Reports. 8 (1): 6340. Bibcode:2018NatSR...8.6340B. doi:10.1038/s41598-018-24724-1. PMC 5910436. PMID 29679057.
- ^ a b Mintz CS (December 1999). "Gene transfer in Legionella pneumophila". Microbes and Infection. 1 (14): 1203–1209. doi:10.1016/s1286-4579(99)00241-5. PMID 10580276.
- ^ Ditommaso, Savina; Giacomuzzi, Monica; Rivera, Susan R. Arauco; Raso, Roberto; Ferrero, Pierangela; Zotti, Carla M. (2014-09-05). "Virulence of Legionella pneumophila strains isolated from hospital water system and healthcare-associated Legionnaires' disease in Northern Italy between 2004 and 2009". BMC Infectious Diseases. 14 (1): 483. doi:10.1186/1471-2334-14-483. ISSN 1471-2334. PMC 4168204. PMID 25190206.
- ^ a b c d e f Steinert, Michael; Heuner, Klaus; Buchrieser, Carmen; Albert-Weissenberger, Christiane; Glöckner, Gernot (2007-11-12). "Legionella pathogenicity: Genome structure, regulatory networks and the host cell response". International Journal of Medical Microbiology. Special issue: Pathogenomics. 297 (7): 577–587. doi:10.1016/j.ijmm.2007.03.009. ISSN 1438-4221. PMID 17467337.
- ^ Burmeister, Alita R. (2015). "Horizontal Gene Transfer: Figure 1". Evolution, Medicine, and Public Health. 2015 (1): 193–194. doi:10.1093/emph/eov018. ISSN 2050-6201. PMC 4536854. PMID 26224621.
- ^ a b Michod RE, Bernstein H, Nedelcu AM (May 2008). "Adaptive value of sex in microbial pathogens". Infection, Genetics and Evolution. 8 (3): 267–285. Bibcode:2008InfGE...8..267M. doi:10.1016/j.meegid.2008.01.002. PMID 18295550.
- ^ a b Charpentier X, Kay E, Schneider D, Shuman HA (March 2011). "Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila". Journal of Bacteriology. 193 (5): 1114–1121. doi:10.1128/JB.01146-10. PMC 3067580. PMID 21169481.
- ^ a b Cunha BA, Burillo A, Bouza E (January 2016). "Legionnaires' disease". Lancet. 387 (10016): 376–385. doi:10.1016/s0140-6736(15)60078-2. PMID 26231463. S2CID 38821013.
